A team of scientists funded by the National Institutes of Health has developed a new tool to monitor under a microscope how cells attach to an adjacent substrate. Studying adhesion events can help researchers understand how tissues grow, how diseases spread, and how stem cells differentiate into more specific cell types. The method provides high-resolution images that can track a cells’ interactions over longer time periods than previously possible.
“Being able to track cells over days with such sensitivity will allow researchers to quantify exactly when these events happen and get a better understanding of the how and why,” says Rosemarie Hunziker, Ph.D., director of the National Institute of Biomedical Imaging and Bioengineering (NIBIB) program in Tissue Engineering and Regenerative Medicine.
Adhesion is a crucial process throughout the cell’s lifespan, from division and differentiation to migration and death. But tracking these events is difficult—whether occurring among cells in the body or the when cells adhere to the surface of a dish in the lab. Most methods rely on expressing fluorescent dyes in the cells, but the fluorescence can be hard to calibrate and exposes cells to fluorescent light for long periods that can damage the cells.
To detect adhesion events, researchers used a photonic crystal sensor, which contains a synthetic crystal capable of manipulating and distinguishing specific wavelengths of light. The sensor was coated with extracellular matrix molecules—proteins that collect near the surface of cells and help promote cell attachment. When a cell attaches to these molecules, the wavelength of light normally refracted off the sensor is shifted, and the sensor registers the shift, detecting the interaction.
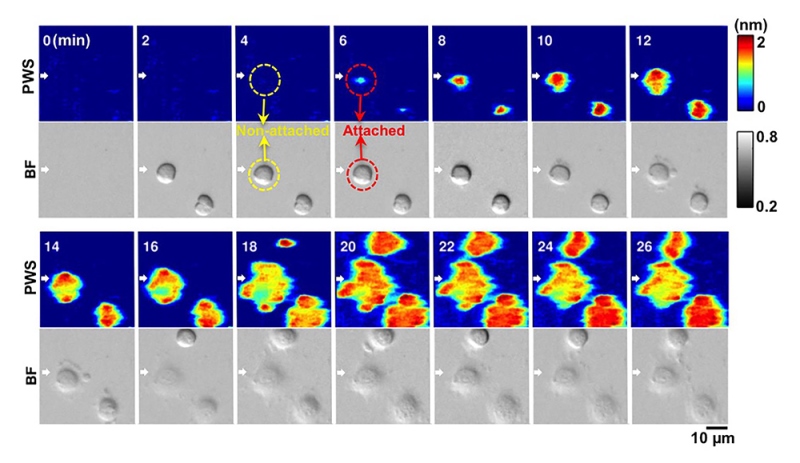
Label-free and dynamic detection of stem-cell adhesion using the photonic crystal-enhanced mircoscope. Credit: Cunningham lab, University of Illinois.
The sensitivity of the system enabled researchers to count and track adhesion events over time. While fluorescent light restricts cell imaging to a few minutes at a time over a span of hours, this system uses a light-emitting diode (LED) source, not fluorescence, so the cells can be imaged over days to weeks without being damaged. The new approach will make it easier to analyze what’s happening at the membrane more reliably and reproducibly, without needing to label the cells.
“We’re addressing this fundamental capability of single cells or multiple single cells within a culture and watching their behavior, their attachment and release behavior over time,” said co-author Brendan Harley, Sc.D., professor of chemical and biomolecular engineering at the University of Illinois at Urbana-Champaign.
Using the crystal sensor in this new way provides many research possibilities. “This is really an untapped imaging approach that hasn’t had a lot of use in looking at cell behavior,” says Harley. “We’re excited about how far can we push this tool to really watch differentiation of stem cells in real time over multiple day processes.”
“All these things that happen through the cell membrane,” says Richard Conroy, Ph.D., former director of the NIBIB program in Molecular Imaging. “The key part here is being able to put some numbers to all of what’s going on.”
Future studies can add drugs and treatments to test their effects on cell binding, use different extracellular matrix molecules to probe the necessary interactions at the surface, or quantify the different cellular receptors involved to better understand how communication at the cell membrane works.
The research was funded in part by NIH grants EB018481 and DK099528.
Quantitative imaging of cell membrane-associated effective mass density using Photonic Crystal Enhanced Microscopy (PCEM). Zhuo Y, Choi JS, Marin T, Yu H, Harley BA, Cunningham BT. Progress in Quantum Electronics. 04 Nov. 2016.
Source: NIBIB